PROTACs and MGDs: The Revolutionary Modalities for Drug Discovery
Stick, Tag, Destroy: The Science of PROTACs and MGDs – For a long time, traditional drug research focused on inhibitors that block the active sites of disease-related proteins. However, around 80% of these proteins remain “undruggable” because they lack clear binding sites.
Traditional drug discovery has long focused on developing inhibitors that block the function of disease-related proteins by occupying their active sites. However, this approach leaves a significant portion of the human proteome, estimated at approximately 80% of disease-related proteins, as “undruggable” due to their lack of distinct active sites or binding pockets1,2. Targeted Protein Degradation (TPD) emerges as a revolutionary alternative, employing small molecules to induce the ubiquitination and subsequent degradation of target proteins via the cell’s natural proteasomal or lysosomal pathways2. Within the TPD landscape, Proteolysis-Targeting Chimeras (PROTACs) and Molecular Glue Degraders (MGDs) stand out as the leading strategies1–3.
PROTACs (Proteolysis-Targeting Chimeras)
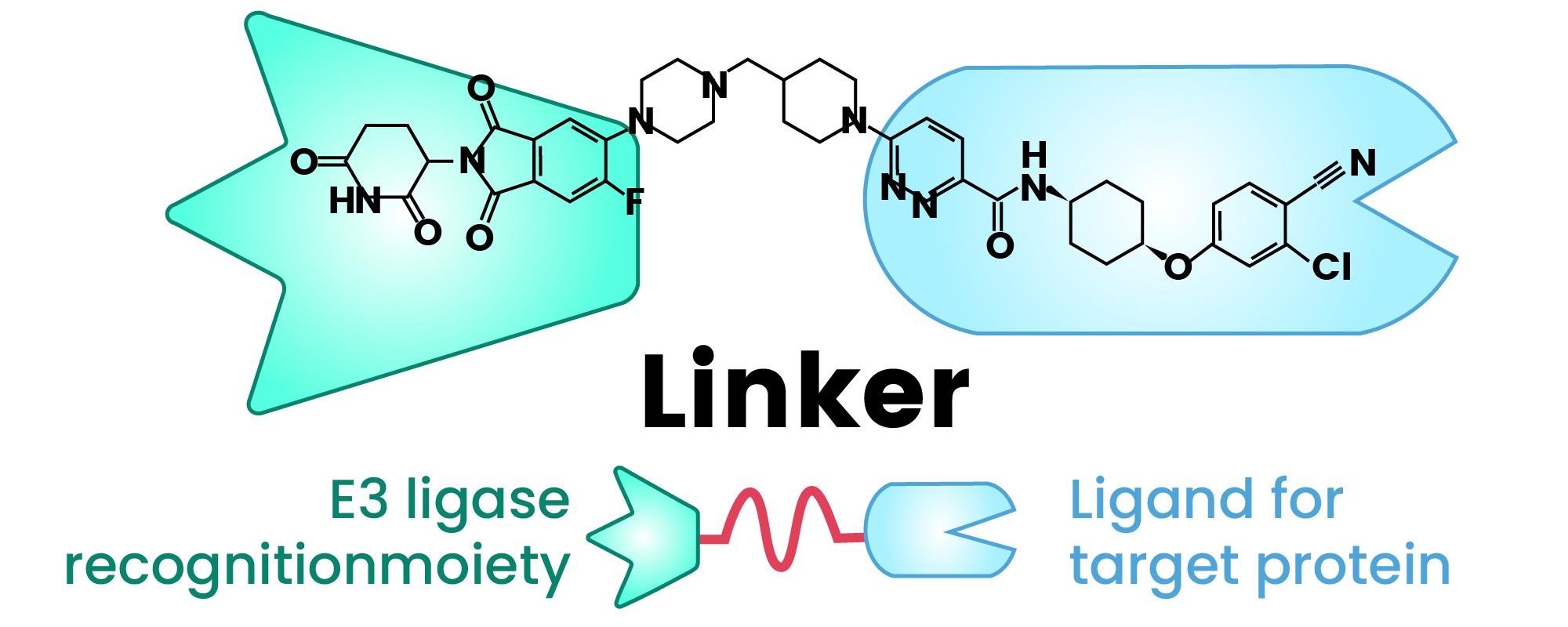
A PROTAC molecule is typically a heterobifunctional compound composed of three essential parts: a ligand that binds to the protein of interest (POI), a ligand that recruits an E3 ubiquitin ligase, and a chemical linker that connects the two (Figure 1)4,5.
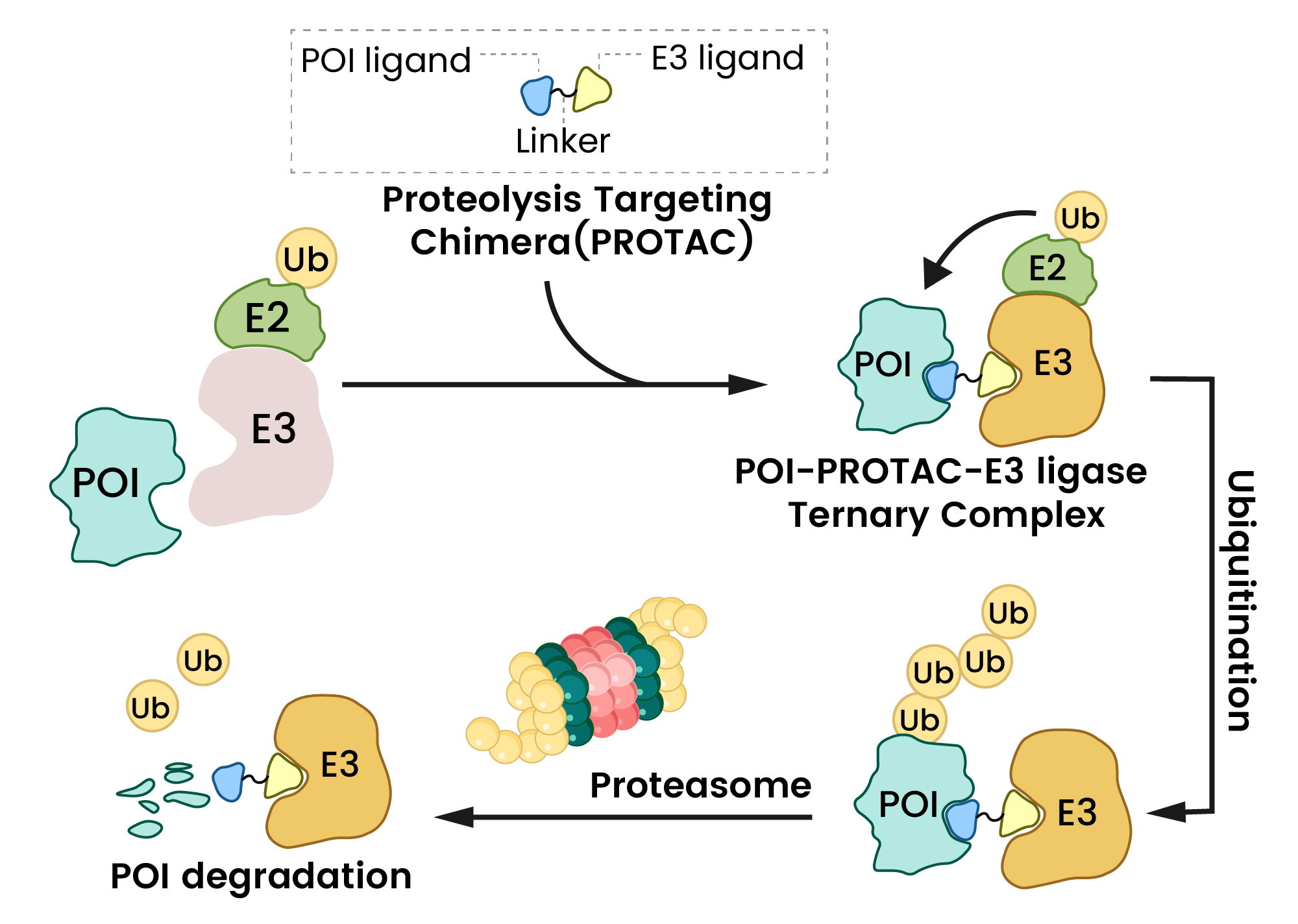
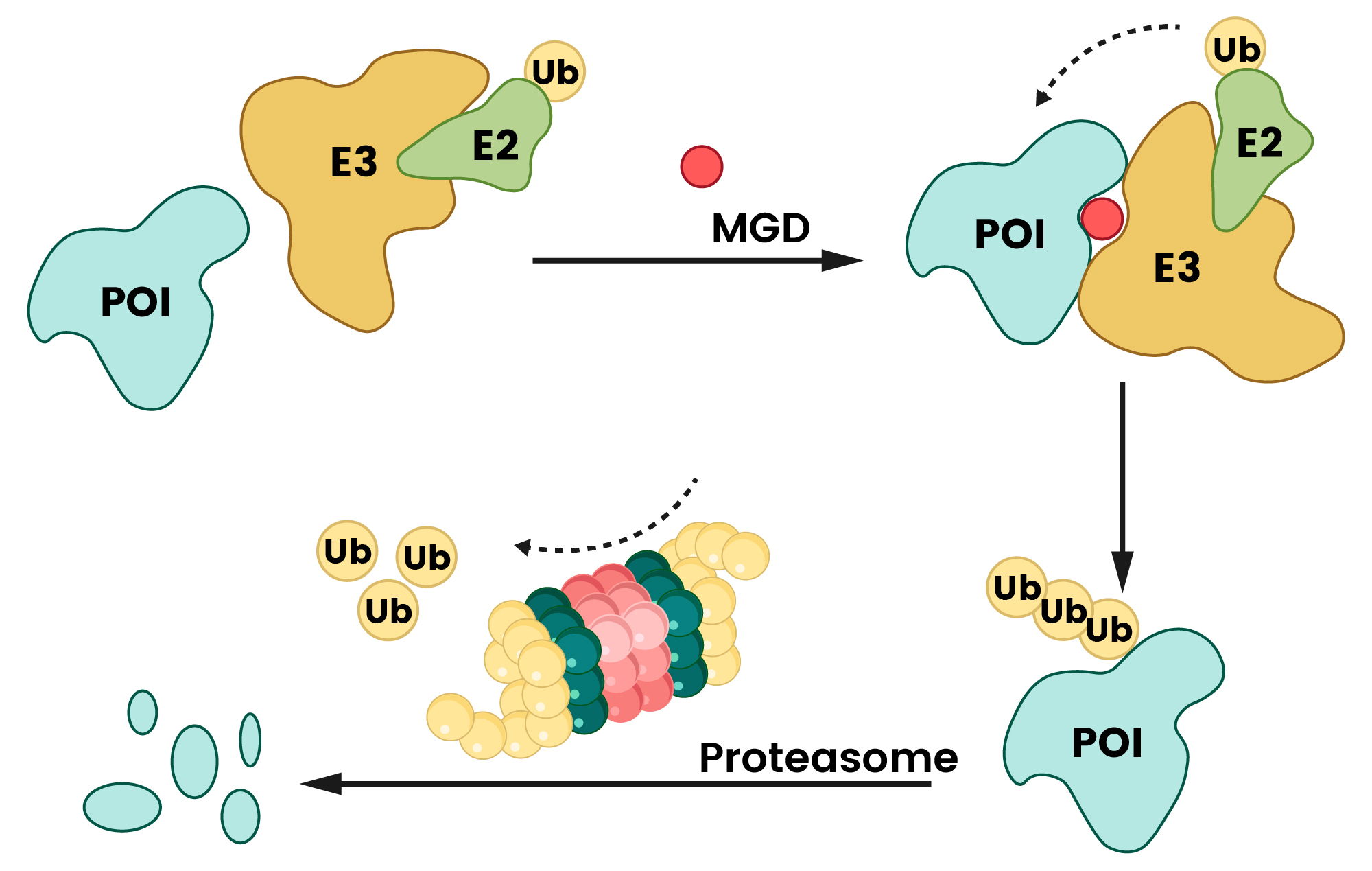
The core of PROTAC action lies in its ability to bring the POI and an E3 ligase into close proximity, forming a ternary complex2,3. This induced proximity facilitates the transfer of ubiquitin tags from the E2 conjugating enzyme (which interacts with the E3 ligase) to the POI2. Once polyubiquitinated, the POI is recognized and rapidly degraded by the 26S proteasome (Figure 2). A key advantage is that after the target protein is degraded, the PROTAC molecule is released and recycled to target another copy of the POI, enabling sustained degradation even at catalytic drug concentrations1,2.
Historical Development and Clinical Progress
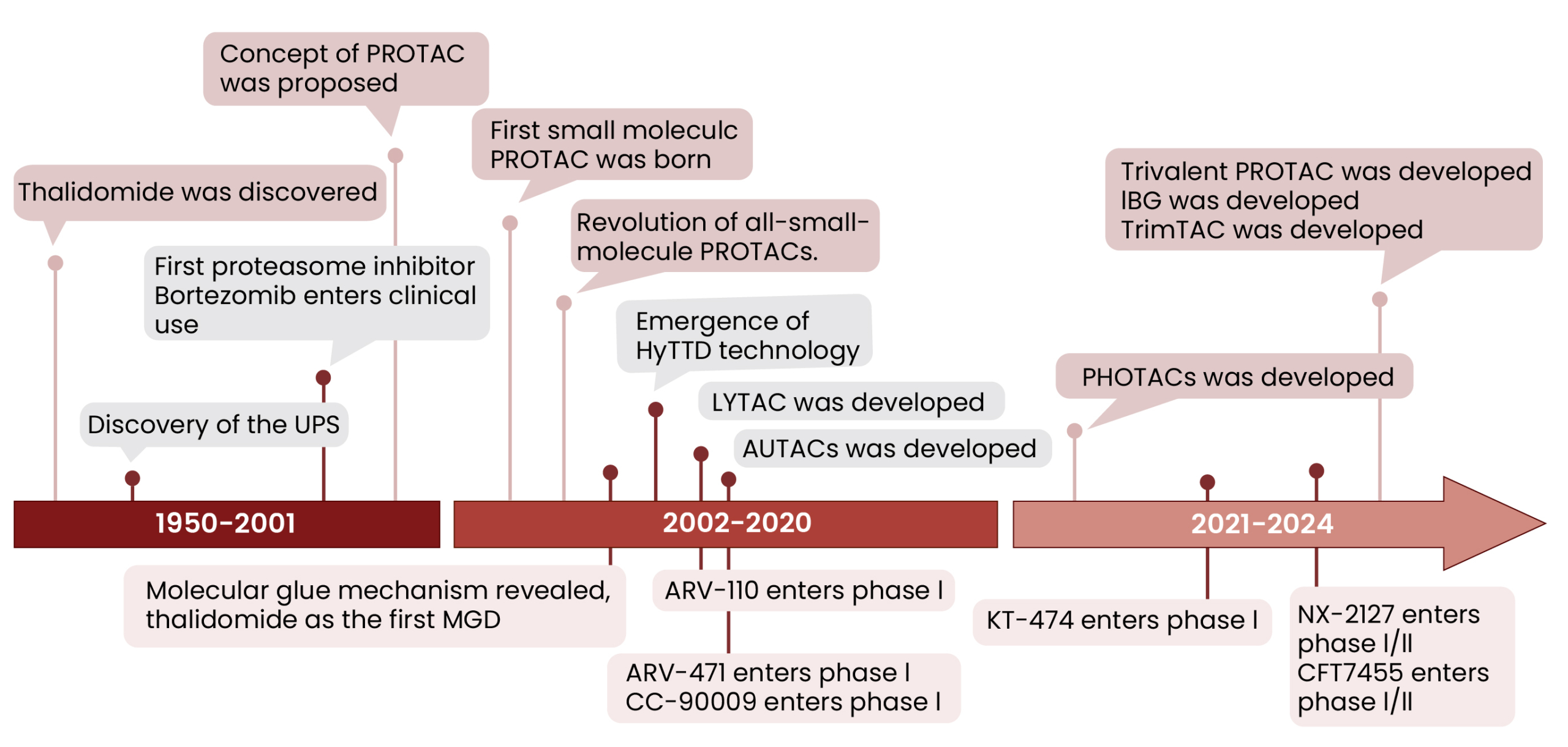
The concept of PROTACs was first introduced in 1999, with the inaugural peptide-based PROTAC molecule (PROTAC-1) developed in 20017. These early PROTACs validated the principle of targeted protein degradation but were hampered by poor cell permeability, limited oral bioavailability, and proteolytic instability. A significant breakthrough occurred in 2008 with the advent of the first fully small-molecule PROTAC (PROTAC 4), which demonstrated superior cell permeability8. This paved the way for identifying small-molecule ligands for key E3 ligases. Cereblon (CRBN) gained prominence in 2010 as the primary target of thalidomide and its analogs. Subsequently, Von-Hippel-Lindau (VHL)-recruiting ligands were discovered in 20121–3. CRBN and VHL are now the most widely utilized E3 ligases in PROTAC design due to their favorable drug-like properties.
Rapid clinical development has followed, with several PROTACs entering clinical trials. Bavdegalutamide (ARV-110), an androgen receptor (AR)-targeting PROTAC, completed Phase II trials for prostate cancer9. Vepdegestrant (ARV-471), targeting the estrogen receptor (ER), has progressed to NDA/BLA for ER+/HER2− breast cancer2,10. NX-2127, a Bruton’s tyrosine kinase (BTK) degrader, has shown efficacy in overcoming drug resistance in B-cell malignancies11. Over 30 heterobifunctional protein degraders are currently under investigation in Phase I-III trials1. Table 1 summarizes representative clinical-stage PROTACs, their target proteins, the E3 ligases they recruit, and their current clinical phases.
Table 1. Representative clinical-stage PROTACs, their target proteins, the E3 ligases they recruit1–3.
| Molecule | Target Protein | E3 Ligase | Clinical Phase |
| ARV-471 | ER | CRBN | NDA/BLA |
| ARV-766 | AR | CRBN | Phase II |
| ARV-110 | AR | CRBN | Phase II |
| DT-2216 | Bcl-XL | VHL | Phase I/II |
| NX-2127 | BTK | CRBN | Phase I |
| NX-5948 | BTK | CRBN | Phase I |
| CFT1946 | BRAFV600 | CRBN | Phase I |
| KT-474 | IRAK4 | CRBN | Phase II |
Challenges and Optimization
Despite their potential, PROTACs face challenges such as their complex structures and high molecular weight (often >700 Da), which frequently places them outside’s “rule of five”. This can lead to poor cell permeability, metabolic instability, and unfavorable pharmacokinetic (PK) profiles2,12. To address these, rational design strategies prioritize linker optimization and E3 ligase ligand optimization13. Incorporating rigid structures like spirocycles or piperidines into the linker can significantly enhance degradation potency and oral bioavailability2. Optimizing E3 ligands involves designing novel scaffolds, such as phenyl glutarimide (PG) derivatives and TX-16, which offer improved stability and affinity compared to traditional thalidomide derivatives2,4,12,13.
Molecular Glue Degraders (MGDs)
Unlike PROTACs’ bivalent architecture, MGDs are monovalent small molecules. They act by inducing or stabilizing novel protein-protein interactions (PPIs) between an E3 ligase and a target protein (POI). This interaction tags the target protein for degradation via the ubiquitin-proteasome pathway2,3. MGDs achieve this by modifying the surface characteristics of substrate receptors and E3 ligases, fostering “non-native” interactions that promote ubiquitination and subsequent degradation3,14. The discovery of MGDs has largely been through serendipitous observation or repurposing existing drug molecules, making their rational design challenging due to the unpredictable nature of these induced interactions15.
Advantages and Challenges
MGDs possess several encouraging features. Their monovalent structure results in simpler molecules with lower molecular weights, which more readily adhere to “Lipinski’s rule of five”. This generally leads to better drug-likeness and favorable pharmacokinetic profiles compared to PROTACs2. Furthermore, MGDs do not necessarily require a binding pocket on the target protein, expanding the range of “undruggable” proteins they can degrade1,2. Thalidomide, lenalidomide, and pomalidomide were among the first MGDs discovered and have been approved for treating erythema nodosum, myelodysplastic syndromes, and multiple myeloma, respectively1,2. Other MGDs, such as CC-90009 (targeting GSPT1) and E7820 (targeting RBM39), are also in clinical development2,3.
The rational design of MGDs remains a significant challenge. Their mechanisms are inherently unpredictable, and clinical translation faces hurdles such as variable degradation efficiencies across tissues, limited predictive biomarkers, and unanticipated off-target effects. Recent efforts in rational MGD design include strategies like introducing covalent handles to existing inhibitors, effectively converting them into degradation-competent MGDs1–3,12,15.
Conclusion and Future Outlook
TPD technologies, particularly PROTACs and MGDs, represent a groundbreaking shift in drug development, offering complete elimination of target proteins rather than mere inhibition. This capacity to degrade “undruggable” proteins provides new therapeutic avenues for various diseases, notably cancers. Despite the remarkable progress, challenges remain, including improving metabolic stability, addressing potential off-target effects, and expanding the array of available E3 ligases beyond the currently dominant CRBN and VHL. The integration of advanced computational methods, such as AI-aided drug design, and comprehensive analytical approaches like multi-omics profiling, is anticipated to accelerate the identification of novel degraders and enhance their selectivity and efficacy2,3. As the field continues to evolve, PROTAC and MGD technologies promise to unlock new therapeutic possibilities for diseases previously considered intractable.
SignalChem Biotech’s Contribution to Targeted Protein Degradation Research
As targeted protein degradation technologies advance, SignalChem Biotech empowers researchers with a robust suite of recombinant E3 ligases—including key molecules like CRBN and VHL—as well as a broad portfolio of target proteins, antibodies, and functional assay reagents. These high-quality, validated proteins and reagents enable the efficient screening, mechanistic interrogation, and optimization of novel PROTACs and molecular glue degraders.
References:
- Pan, M., Fu, Z., Hou, H., Yang, C. & Li, J. Proteolysis-Targeting Chimera (PROTAC): A Revolutionary Tool for Chemical Biology Research. Small Methods Preprint at https://doi.org/10.1002/smtd.202500402 (2025).
- Pan, Y., Wang, Y. & Gou, S. Proteolysis targeting chimera, molecular glue degrader and hydrophobic tag tethering degrader for targeted protein degradation: Mechanisms, strategies and application. Bioorg Chem161, 108491 (2025).
- Zhang, B. et al. Rational Design of Dual Degraders by Incorporating Molecular Glue Structural Features into PROTAC Degraders. J Med Chem68, 10268–10298 (2025).
- Ito, T. Protein degraders – from thalidomide to new PROTACs. J Biochem175, 507–519 (2024).
- Rutherford, K. A. & McManus, K. J. PROTACs: Current and Future Potential as a Precision Medicine Strategy to Combat Cancer. Mol Cancer Ther23, 454–463 (2024).
- Pagan, J. et al. Role of the Ubiquitin-Proteasome System in the Heart. Circulation Research112, 1046–1058 (2013).
- 7. Sakamoto, K. M. et al.Protacs: Chimeric Molecules That Target Proteins to the Skp1-Cullin-F Box Complex for Ubiquitination and Degradation. https://www.pnas.org (2001).
- 8. Schneekloth, A. R., Pucheault, M., Tae, H. S. & Crews, C. M. Targeted intracellular protein degradation induced by a small molecule: En route to chemical proteomics. Bioorg Med Chem Lett18, 5904–5908 (2008).
- 9. Chirnomas, D., Hornberger, K. R. & Crews, C. M. Protein degraders enter the clinic – a new approach to cancer therapy. Nat Rev Clin Oncol20, 265–278 (2023).
- 10. Hamilton, E. P. et al. ARV-471, an estrogen receptor (ER) PROTAC degrader, combined with palbociclib in advanced ER+/human epidermal growth factor receptor 2–negative (HER2-) breast cancer: Phase 1b cohort (part C) of a phase 1/2 study. Journal of Clinical Oncology40, TPS1120–TPS1120 (2022).
- 11. Robbins, D. W. et al. Discovery and Preclinical Pharmacology of NX-2127, an Orally Bioavailable Degrader of Bruton’s Tyrosine Kinase with Immunomodulatory Activity for the Treatment of Patients with B Cell Malignancies. J Med Chem67, 2321–2336 (2024).
- 12. Pu, C. et al. Current strategies for improving limitations of proteolysis targeting chimeras. Chinese Chemical Letters34, 107927 (2023).
- 13. Zagidullin, A., Milyukov, V., Rizvanov, A. & Bulatov, E. Novel approaches for the rational design of PROTAC linkers. Open Exploration 2019 1:51, 381–390 (2020).
- 14. Malone, M. L., Sanchez, N. A., Hu, S. L. & Phelps, C. B. Application of DELs for E3 Ligase Ligand Discovery and Targeted Protein Degradation. DNA-encoded Library Technology for Drug Discovery 134–156 (2025) doi:10.1039/9781788016032-00134.
- 15. Liu, Y., Bai, J., Li, D. & Cang, Y. Routes to molecular glue degrader discovery. Trends Biochem Sci50, 134–142 (2025).


 Unsplash+
Unsplash+ Unsplash+
Unsplash+ FIDA Biosystems ApS
FIDA Biosystems ApS