New technology speeds up mapping of DNA-protein interaction
Scientists at EPFL have developed a technique that can be a game-changer for genetics by making the characterisation of DNA-binding proteins much faster, more accurate, and efficient.
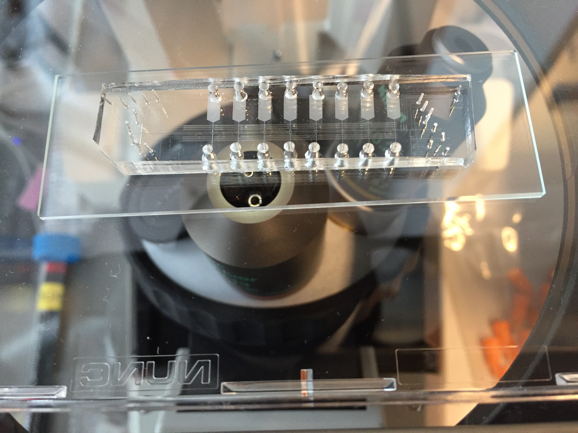
Researchers at the École polytechnique fédérale de Lausanne (EPFL) have developed a semiautomated microfluidics-based technique that can greatly speed up the identifcation of the 1,300-2,000 transcription factors, many of which combine with others into heterodimers in order to bind genes and induce their transcription into RNA. A patent has been filed and the group headed by Bart Deplancke aims to commercialise the findings. Right now, we are exploring different options, Deplancke told European Biotechnology. However, no decision on the commercialisation path has been made. Up to now, the researchers have determined the DNA binding properties of more than 60 transcription factors from humans and model organisms including nine new ones (Nature Methods, doi: 10.1038/nmeth.4143). The process cuts the time for transcription factor identification from days to less than an hour.
The new technique called SMiLE-seq (semiautomated protein-dnA interaction characterisation technology, selective microfluidics-based ligand enrichment followed by sequencing) makes use of microfluidics and works by attaching small amounts of the transcription factor in a chip with micrometer-size channels that allow liquid to flow through. Once the transcription factors are attached to the chip’s surface, a large library of random DNA is gently pumped into the chip and flows over them. This allows the transcription factors to recognise their corresponding DNA sequences. Thereafter, the transcription factor-DNA complex is physically trapped by dropping down a microfluidically steered button, while the DNA that is not bound is simply washed away. Next, the bound DNA is taken off the device and prepared for sequencing to identify which part of it got caught by the transcription factors. This information is fed into a software that helps to better predict their in vivo DNA-binding profiles. According to the authors, SMiLE-seq is not limited by either the length of the DNA target sequence, nor is it biased toward stronger affinity protein-DNA interactions as current methods might be.
According to the publication, Deplancke’s team used SMiLE-seq to determine 58 full-length human, mouse, and Drosophila transcription factors, successfully analysing several that were never studied before. In a press release, however, the EFPL spoke of 67 factors analysed. In the future, he intends to exploit the technique’s versatility into other molecules, such as RNA. A patent has been filed. However, according to Deplancke it is not yet clear what the best option for commercialisation is.



 Vetter Pharma International GmbH
Vetter Pharma International GmbH HeiQ/AeoniQ, Renewcell
HeiQ/AeoniQ, Renewcell