Researchers reconstruct SARS-CoV-2 evolution
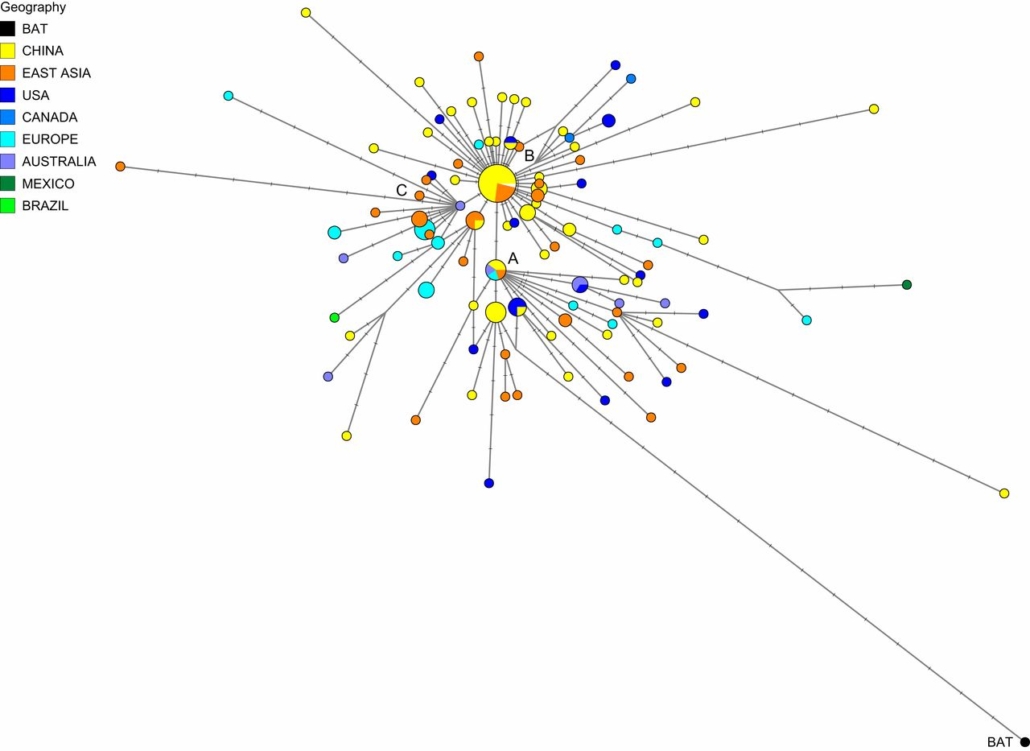
British researchers have tabled a phylogenetic tree showing different subtypes of SARS-CoV-2 developed in Asia, Europe and the US.
The work published by Peter Forster and collegues from University Cambridge in PNAS is based on a phylogenetic network of the about almost completely sequenced 160 SARS-CoV-2 genomes sampled from across the world and stored in the GISAID database, which contained a compilation of 253 severe SARS-CoV-2 genomes. The researchers applied a technique to reconstruct language prehistory to analyse the spread of the SARS-CoV-2 virus.
Comparative analyses of character-based phylogenetic networks suggest that all genomes are closely related and under evolutionary selection in their human hosts, sometimes with parallel evolution events, that is, the same virus mutation emerges in two different human hosts.
According to the phylogenetic network analysis, they identified three central variants they named A, B, and C and which have characteristic amino acid changes. A is the ancestral type according to the bat outgroup coronavirus. The A and C types are found in significant proportions outside East Asia, that is, in Europeans and Americans. In contrast, the B type is the most common type in East Asia, and its ancestral genome appears not to have spread outside East Asia without first mutating into derived B types, pointing to founder effects or immunological or environmental resistance against this type outside Asia.
The network faithfully traces routes of infections for documented coronavirus disease 2019 (COVID-19) cases, indicating that phylogenetic networks can likewise be successfully used to help trace undocumented COVID-19 infection sources, which can then be quarantined to prevent recurrent spread of the disease worldwide.



 Unsplash+
Unsplash+