Researchers unravel how ADEP antibiotics work
German researchers have determined the mechanism of a ADEPs, a novel class of antibiotics, allowing for pharmacological optimisation.
Fighting antimicrobial resistance has been named one of the prioritary goals by the World Health Organization (WHO). However, as the number of bacterial pathogens resistant even to last resort antibiotics is growing, governments and initiatives across the world have invested much money to find novel antibiotic mechanisms of action that help overcome bacterial resistance for a while. A German research team has now made a major step to develop such a new class of antibiotics.
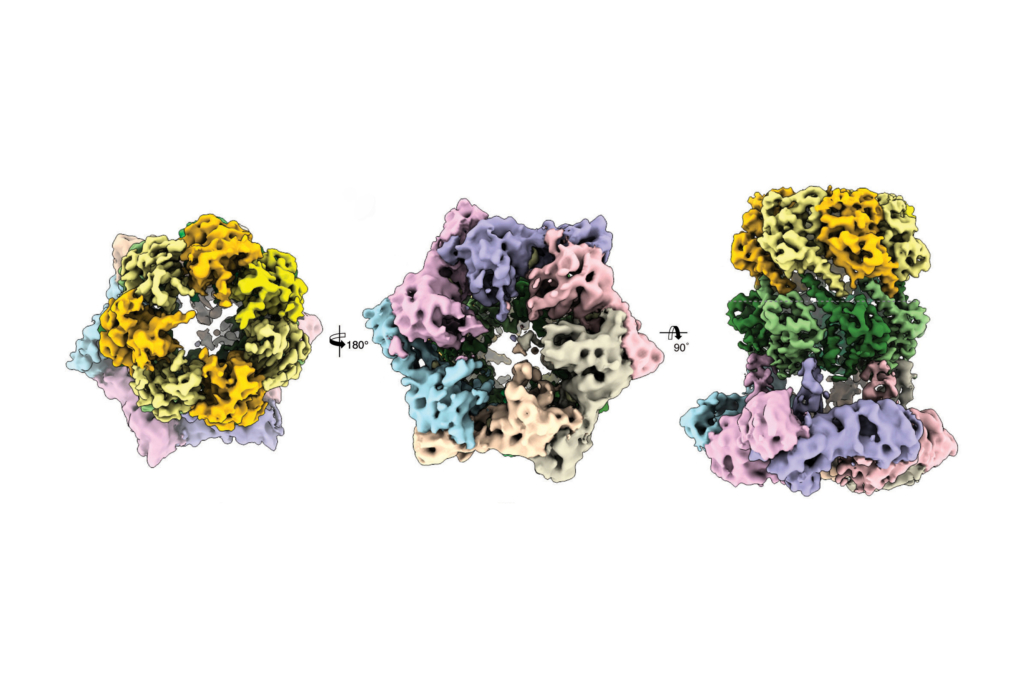
Using cryogenic electron microscopy, researchers led by Stefan Raunser at MPI for Raunser, from the Max Planck Institute of Molecular Physiology in Dortmund, and Stefan Sieber at LMU Munich found out how so-called acyldepsipeptide (ADEP) antibiotics can be optimised to kill even multiresistant pathogens. In Nature Structural & Molecular Biology, the researchers report how ADEPs switch the activity of the bacterial protease ClpP so that it degraded all proteins of the bacterial pathogen Listeria monocytogenes in an uncontrolled manner.
Usually, bacterial ClpP forms a complex with the protein ClpX, which recognises defective proteins and this way labels them for targeted degradation. Specifically ClpX utilises the energy of ATP binding and hydrolysis to engage, unfold and translocate defective proteins into the catalytic chamber of tetradecameric ClpP protein, which specificalls degrades them.
Solving the structure of the ClpP/ClpX complex, the researchers found that acyldepsipeptides (ADEP) bind on the same spot at ClpP protease as ClpX but in a different manner. Whereas ClpX does not lead to an alteration in the structure of ClpP, ADEP brings about an unintended opening of the complex. As a result, intact proteins are also degraded in an uncontrolled manner and independently from ClpX.
The clarification of this mechanism is a milestone on the way to the development of innovative antibiotic substances targeting ClpP. According to Sieber and Raunser, this mechanism of action has considerable innovation potential in the fight against pathogenic bacteria. Whereas common antibiotics act through the inhibition of vital processes, in this case the antibacterial effect is achieved through the activation of a process, they said.
Furthermore, besides controlled degradation of defective proteins, ClpP plays an important role in bacterial metabolism. The enzyme acts also as a regulator in the production of an arsenal of bacterial toxins which are primarily responsible for the pathogenic effect of many pathogens. Sieber’s group has developed diverse potent inhibitors against ClpP and ClpX which stop the production of bacterial toxins and can therefore more or less disarm them.


 chokniti - Adobe Stock
chokniti - Adobe Stock