Roche unveils completely new sequencing approach
Roche researchers have introduced a greatly improved long-read nanopore sequencing method that has the potential to erase the sequencing-by-synthesis approach of market leader Illumina.
Researchers at Roche have introduced a greatly improved nanopore sequencing method that has the potential to revolutionise next-generation sequencing . The scalable and flexible ‘Sequencing by Expansion’ (SBX) promises to speed up genome sequencing enormously which might contribute to multiple biomedical applications as called for in a position paper “The untapped potential of Comprehensive Genomic Profiling” published today by ECGP and EUCOPE.
According to the European Coalition for Comprehensive Genomic Profiling (ECGP) and the pharma federation EUCOPE, access to comprehensive genomic profiling in Europe is still limited, blocking routine testing for all relevant cancer mutations and the rapid adoption of personalised medicine in Europe..
In contrast to Illumina’s sequencing standard Sequencing by Synthesis (SBS), Roche’s new chemical process is based on the expansion of target nucleic acids passing through a nanopore (10.1101/2025.02.19.639056v1). The individual bases are expanded into surrogate polymers, so-called Xpandomers. These reporter molecules are many times longer than the original nucleic acid sequence and therefore generate particularly strong signals with minimal background noise – the biggest problem with other nanopore sequencing approaches to date.
According to Roche authors Mitu Chadhary and Mark Kokoris, the SBS approach enables particularly fast, accurate and flexible determination of the DNA sequence. ‘The speed and accuracy of SBX has the potential to revolutionise the application of sequencing in research and healthcare,’ said Matt Sause, CEO of Roche Diagnostics, adding that the technology is suitable for a wide range of genome, exome and RNA sequencing applications. It can sequence up to seven complete genomes per hour at 30x coverage, which equates to 5 billion duplex reads per hour – in both strands of DNA sequenced simultaneously.
Two key new developments
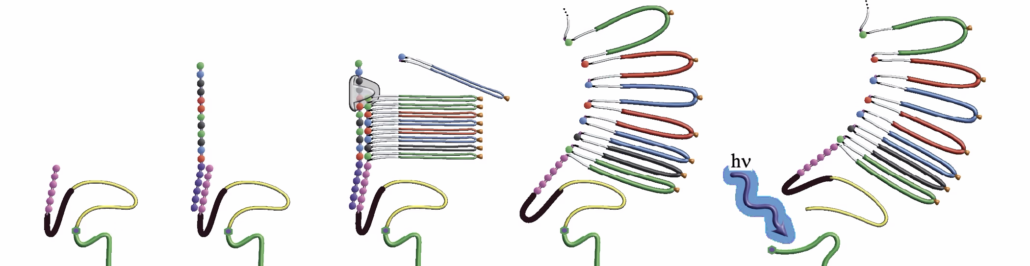
The first innovation is the SBX expansion approach developed by Mark Kokoris and Robert McRuer at Stratos Genomics, which Roche acquired in 2020. The aim was to overcome the limitations of previous methods, which lie in the dense packing of nucleic acid sequences and lead to a poor signal-to-noise ratio. Solving the signal-to-noise problem is a key efficiency factor for this technology. “Thanks to this capability, we can work flexibly with one and the same sequencing system across different throughput levels, which offers users a significant advantage,” says Kokoris, now head of SBX technology at Roche, adding that the sequences are first expanded in xpandomers for subsequent nanopore sequencing.
The basis for this are expandable nucleoside triphosphates, the X-NTPs, which contain highly significant reporter codes. A specially developed polymerase incorporates the X-NTP molecules into the growing Xpandomer – stabilised by special polymerase enhancers (PEMs). After synthesis, the bonds in the Xpandomer are cleaved, resulting in a long molecule that is around fifty times longer than the original sequence.
The second major innovation concerns the read-out of the sequences. After synthesis, the Xpandomer is guided over a nanopore where voltage pulses control its movement and the reporter codes are read out with high precision. This is done using a high-throughput sensor module developed by Genia Technologies, which is based on complementary metal oxide semiconductor (CMOS) technology and is designed to enable ultra-fast and parallel processing.
Specifically, each of the four, easily differentiated X-NTPs (one for each base), act as substrates for template-dependent, polymerase-based replication. The polymerase, XP synthase, has been carefully engineered to incorporate large X-NTP monomers, enabling >99.3% mean raw read accuracy, uniform GC coverage, and longer read lengths. Polymerase enhancing moieties, or PEMs, are also added to the synthesis reaction to assist the polymerase in properly incorporating X-NTPs into the growing polymer. By stabilising the extending molecule, PEMs play an important role in increasing read length beyond traditional short-read sequencing technologies. Following synthesis of the surrogate molecule, acid-cleavable bonds are broken, allowing the newly synthesized Xpandomer to extend 50X longer than the original DNA molecule.
The Xpandomer molecule is then routed through a biological nanopore in a highly efficient and accurate manner. Movement of the Xpandomer through the pore is guided by voltage pulses that advance the Xpandomer through the pore one reporter code at a time. The highly differentiated reporter codes are easily measured during this translocation process via a scalable complementary metal oxide semiconductor (CMOS)-based array, which combines electrodes, detection circuits, and analog-to-digital conversion. Because the CMOS array contains roughly eight million microwells (each containing a nanopore), measurement occurs in a massively parallel, highly controlled manner without the convolution issues of traditional nanopore sequencing. The result is the cost-effective measurement of hundreds of millions of bases per second, bypassing the traditional approach of cyclical incorporation and measurement of a single base at a time.


 Araris Biotech AG
Araris Biotech AG Roche
Roche Qiagen
Qiagen