Scientists map ulcerative colitis
Scientists from Leuven, Rotterdam and San Diego have published a sequencing-based map of ulcerative colitis paving the way to new treatments.
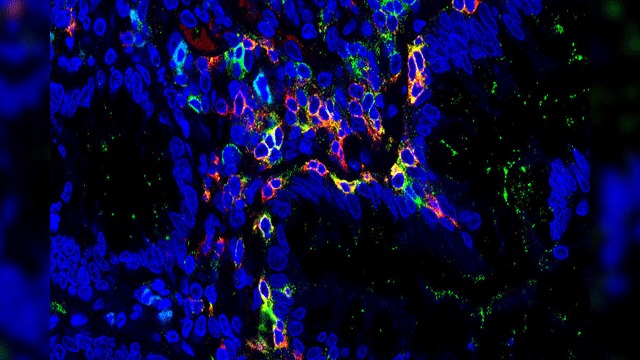
The study that combines multiple next-generation genetic sequencing technologies extensively characterize the diverse array of cells and cellular components that contribute to ulcerative colitis, providing a cellular atlas that could help inform future treatment of the chronic inflammatory disease. According to the authors under Danny Huylebroeck, each subset of T and B cells identified, may serve as a useful resource to inform future therapies for colitis and other inflammatory bowel disorders (IBDs) such as Crohn’s disease.
Using a combination of cell sorting and single-cell T and B cell receptor analysis, the research team identified four different sub-subsets of CD8+ TRM cells in both healthy patients and those with colitis. In the latter cohort, these TRM cells exhibited a greater tendency to differentiate into an inflammatory state, associated with increased expression of the transcription factor Eomesodermin (Eomes). The authors hypothesise that Eomes may be a critical regulator of inflammatory TRM cells during ulcerative colitis, and therefore could serve as a promising target for future IBD research.
In 2019, researchers from University of Erlangen reported that TRM cells might cause flare-ups in chronic inflammatory diseases such as Morbus Crohn or ulcerative colitis.



 adobe.stock.com - ipopba
adobe.stock.com - ipopba BioDlink
BioDlink