S-Protein: Glycosylation pattern revealed
British researchers have deciphered the glycosylation pattern of the viral spike protein of SARS-CoV-2 laying the foundation for vaccine development.
Using high-resolution mass spectrometry, researchers at the University of Southampton have mapped glycan-processing states of the spike protein complex that allows the SARS-CoV-2 virus to infect human cells – finding that SARS-CoV-2 S glycans differ from typical host glycan processin. This may have implications in vaccine design.
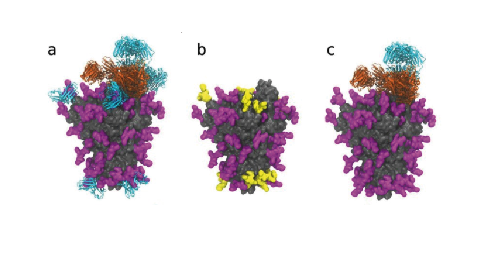
As scientists seek to combat the virus that causes COVID-19, 99% of the efforts to develop a vaccine have focused on the spike, a protein complex composed of three protomers that protrudes from the virus and binds to the ACE2 receptor on the surfaces of human cells, or its receptor-binding region. Each protomer harbors 22 chemical sites that can undergo glycosylation. How these sites are glycosylated are beleived to affect, which cells the virus can infect.
Glycans could also shield some regions on the spike from being neutralised by antibodies. Seeking insight, Yasunori Watanabe et al. expressed and purified recombinant glycosylated spike complexes, then used enzymes to cut them into peptides each containing a single glycan but representing all glycan sites. The researchers then used mass spectrometry to determine the glycan composition at each site.
They report that the SARS-CoV-2 S protein is less densely glycosylated than some other viral glycoproteins, possessing a sparse glycan shield, which may be beneficial for the elicitation of potent neutralizing antibodies. Their analysis provides a benchmark that can be used to measure the quality of the spike antigen as researchers and companies develop new vaccines and antibody tests.



 adobe.stock.com - ipopba
adobe.stock.com - ipopba BioDlink
BioDlink