Sleeping sickness target deciphered
Researchers at the University of Luebeck have revealed the structure of the IMP dehydrogenase, a central target of T. brucei, which causes sleeping sickness.
Using ultra-bright X-ray flashes, a team of researchers led by Lars Redecke from the University of Lübeck and DESY, Christian Betzel from the University of Hamburg, and Henry Chapman from DESY solved the protein structure of a drug target against sleeping sickness. In Nature Communications, the scientists reported the the detailed spatial structure of IMP dehydrogenase, a vital enzyme of the parasite Trypanosoma brucei. It provides a possible blueprint for a drug that specifically blocks the enzyme thereby killing the parasite.
Sleeping sickness is transmitted by the bite of tsetse flies, which is endemic in tropical Africa. In the body, the parasite first multiplies under the skin, in the blood and in the lymphatic system and then migrates to the central nervous system. If left untreated, the disease ends fatal. However, intensive control measures have helped to decrease the number of registered cases in recent years. Nevertheless, sleeping sickness is still considered one of the most important tropical diseases with 65 million people living in the sub-Saharan risk area. War, displacement and migration can cause the disease to flare up.
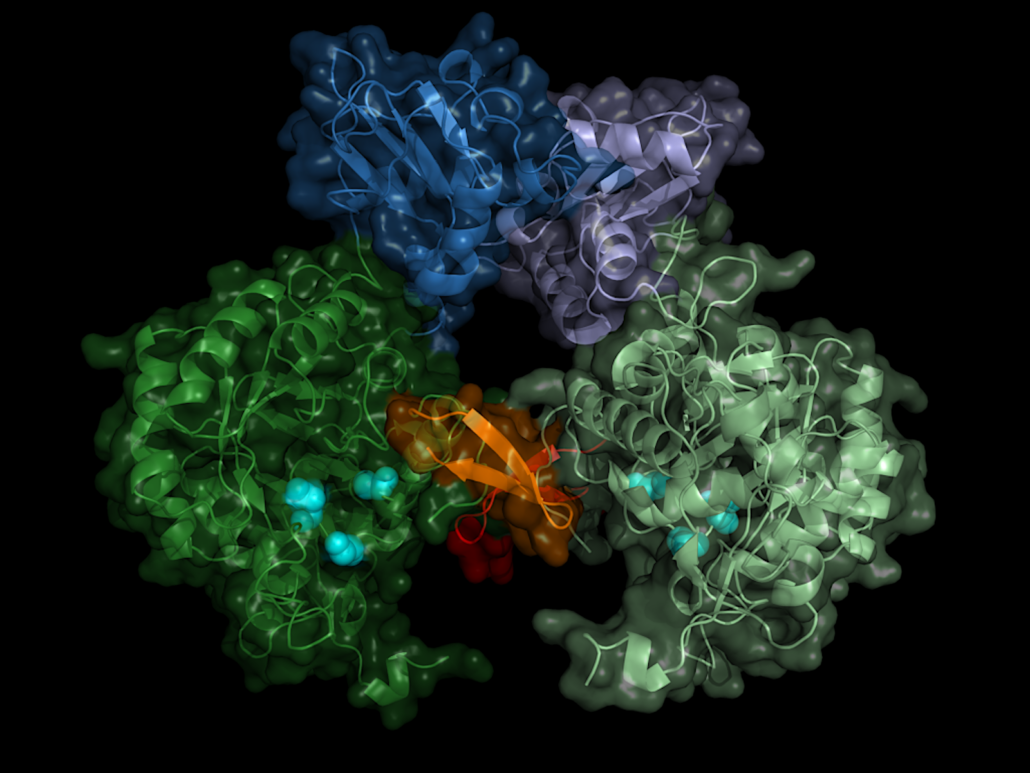
Searching for a starting point for drugs against the pathogen, the researchers focussed on a central enzyme of the pathogen, inosine-5′-monophosphate dehydrogenase (IMPDH). This enzyme belongs to the central inventory of every organism and is an interesting drug target because it regulates the concentration of guanosine diphosphate and guanosine triphosphate, says Redecke. which the cell needs to supply energy and to build up the genome. If you interrupt this cycle, the cell dies.
The enzyme has a kind of on/off switch that is activated by binding of the cell’s own molecules. A promising approach is to block this switch with a precisely tailored molecule. In order to construct such an inhibitor, the exact spatial structure of the switch must be known. Although the structures of numerous IMP dehydrogenases from other organisms were already known, there had been no success in growing crystals of the Trypanosoma brucei version of the enzyme so far.
The structure biologists therefore took an alternative route, the crystallization of proteins in living insect cells, so-called in cellulo crystallization. The 2.8Å-resolved structure does not only show the exact structure of the enzyme switch, the Bateman region, but also which molecules of the cell activate the switch and how these so-called co-factors dock to the enzyme. The switch is operated by the molecules adenosine triphosphate (ATP) and guanosine monophosphate (GMP).
The data might provide an approach for blocking the parasite’s but not the human IMP dehydrogenase.




 Qiagen
Qiagen